ClustEval
clustering evaluation framework
ClustEval
clustering evaluation framework
If you need additional information about one of the datasets, click on its name and you will be redirected to a page with further details including an overview, how well different clustering methods performed on this dataset.
Available datasets
Gene Expression
bone_marrow (Details)
This data set has been made publicly available by the Broad Institute. It contains microarray gene expression levels of 999 genes for 38 samples of leukemia patients suffering from three different subtypes of acute leukemia. A gold standard is provided consisting of three cancer subtypes.
Publication: Stefano Monti, Pablo Tamayo, Jill Mesirov, Todd Golub. Consensus Clustering: A Resampling-Based Method for Class Discovery and Visualization of Gene Expression Microarray Data. Machine Learning, Volume 52 Issue 1-2, July-August 2003 Pages 91-118, 2003 (Link)bone_marrow_fixLabels (Details)
tcga (Details)
The Cancer Genome Atlas (TCGA) is a project maintaining a database storing molecular information of cancer cells, including gene expression, DNA methylation or copy number aberration. It includes data for many different cancer types, allowing their comparison on a molecular level. A data set has been derived integrating gene expression levels, DNA methylation and copy number aberration of the three different cancer types, namely Breast Invasive Carcinoma (BRCA, 207 samples), Glioblastoma Multiforme (GBM, 67 samples) and Lung Squamous Cell Carcinoma (LUSC, 19 samples). For each type of molecular information the authors calculated pairwise similarities between the samples using Spearman correlation. This resulted in three similarities for every pair of samples, which were then combined by taking their arithmetic mean.
Publication: Nora Speicher. Towards the identification of cancer subtypes by integrative clustering of molecular data. Masters thesis, Saarland University, December 2012. (Link)
Protein Sequence Similarity
astral1_161 (Details)
ASTRAL is a database containing classifications of proteins partially derived from the SCOP database. Different releases of ASTRAL are available, containing data evolved over time. We included the data set ASTRAL95_1_161 derived from ASTRAL as described in. This data set contains pairwise protein similarities generated by applying a similarity function to the blasted proteins contained in the above protein sets. The gold standard to the dataset contains the SCOP classification of each protein.
Publication: Chandonia JM, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL compendium in 2004. Nucleic Acids Research, 32:D189-D192, 2004 (Link)astral_40_seqsim_beh (Details)
brown (Details)
The data set contains pairwise similarities of blasted sequences of 232 proteins belonging to the amidohydrolase superfamily. A gold standard is provided describing families within the given superfamily. According to the gold standard the amidrohydrolase superfamily contains 29 families.
Publication: Shoshana D Brown, John A Gerlt, Jennifer L Seffernick, and Patricia C Babbitt. A gold standard set of mechanistically diverse enzyme superfamilies. Genome Biol, 7(1):R8, 2006. (Link)
Protein Structure Similarity
astral_40_strsim (Details)
Protein-Protein-Interaction
ppi_mips (Details)
The MIPS Mammalian Protein-Protein Database is a database for protein-protein interactions of mammalian species. We used the data set proposed in consisting of a subset of 220 protein complexes of 1562 proteins.
Publication: Philipp Pagel, Stefan Kovac, Matthias Oesterheld, Barbara Brauner, Irmtraud Dunger-Kaltenbach, Goar Frishman, Corinna Montrone, Pekka Mark, Volker Stuempflen, Hans-Werner Mewes, Andreas Ruepp and Dmitrij Frishman. The MIPS mammalian protein-protein interaction database. Bioinformatics, 21(6):832-834, 2005 (Link)
Social Network
zachary (Details)
The data set contains similarities between 34 members of a karate club. The karate club split up into two groups, which is used as a gold standard.
Publication: Wayne W. Zachary. An information flow model for conflict and fission in small groups. Journal of Anthropological Research, 1977 (Link)
Synthetic
chang_pathbased (Details)
gionis_aggregation (Details)
synthetic_cassini (Details)
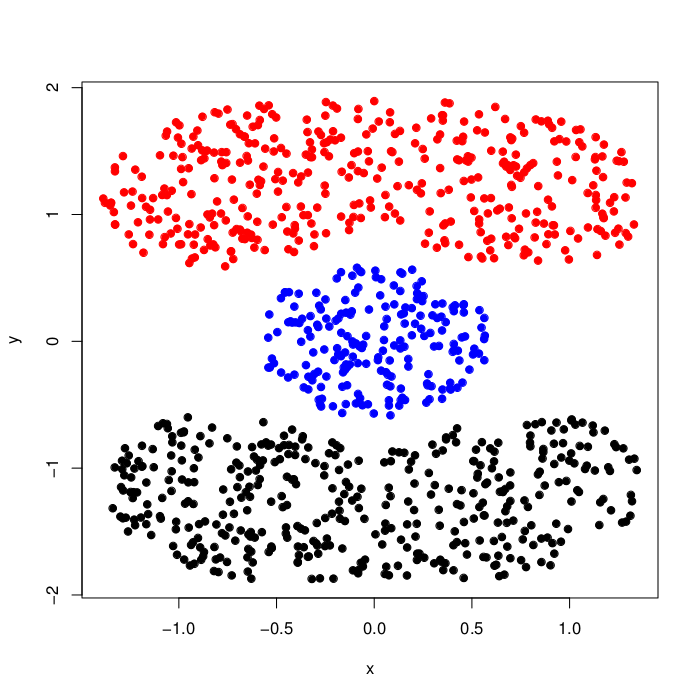 The cassini data set consists of three groups, two banana-shaped clusters (non-convex) bending around a circular cluster in between them.
The cassini data set consists of three groups, two banana-shaped clusters (non-convex) bending around a circular cluster in between them.
synthetic_cuboid (Details)
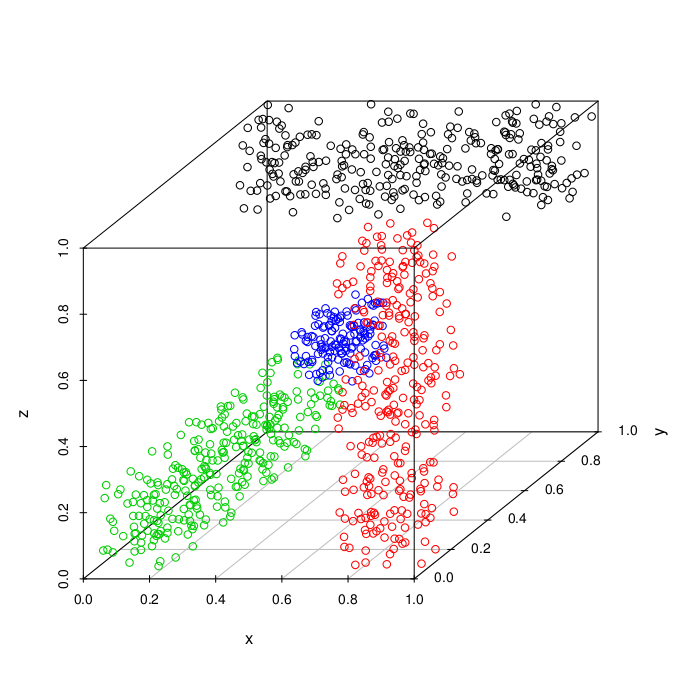 The cuboid data set consists 3 cuboids and one cube in between in a three dimensional space. The cuboids are placed at the edges of a boundary cube.
The cuboid data set consists 3 cuboids and one cube in between in a three dimensional space. The cuboids are placed at the edges of a boundary cube.
synthetic_spirals (Details)
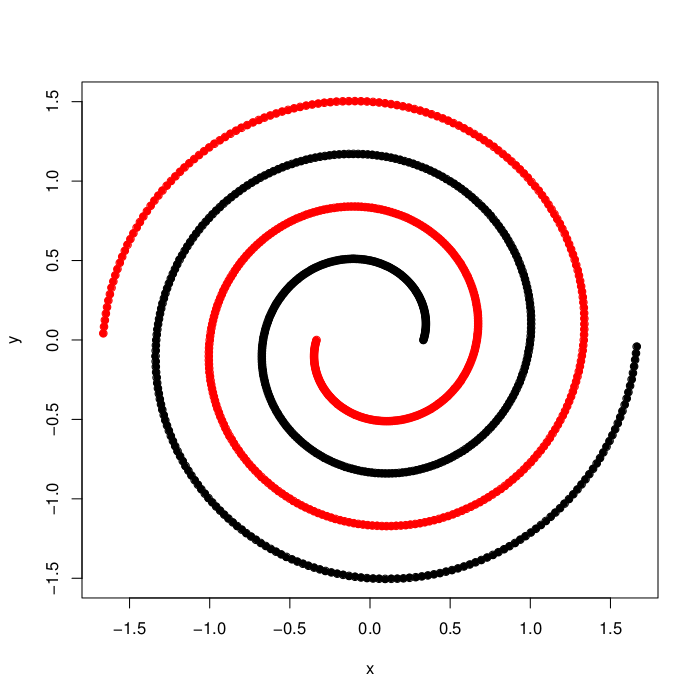 This synthetic data set consists of 250 two-dimensional data objects, distributed over two entangled spirals. Since the data set is non-convex, it is almost impossible to resolve the natural grouping for clustering methods, that are solely based on an approach maximizing between-cluster distances and minimizing within-cluster distances in the original input space. Therefore, methods that either transform the data into a more suitable space, like Spectral Clustering, or that base their cluster identification on local neighborhood similarities will be able to identify the cluster structure.
This synthetic data set consists of 250 two-dimensional data objects, distributed over two entangled spirals. Since the data set is non-convex, it is almost impossible to resolve the natural grouping for clustering methods, that are solely based on an approach maximizing between-cluster distances and minimizing within-cluster distances in the original input space. Therefore, methods that either transform the data into a more suitable space, like Spectral Clustering, or that base their cluster identification on local neighborhood similarities will be able to identify the cluster structure.
twonorm_100d (Details)
twonorm_50d (Details)
veenman_r15 (Details)
zahn_compound (Details)
Word Sense Disambiguation
coli_find (Details)
Text corpora contain occurrences of the same word in different contexts or senses. These texts can be analyzed automatically and word sense disambiguation can be employed to infer the context solely based on the contained words. For this purpose, pairwise similarities between single word occurrences can be calculated by masking and comparing word neighborhoods. Based on these similarities the occurrences can be clustered into potential contexts. This approach has been applied to a text corpus containing occurrences of the word "find".
coli_need (Details)
Text corpora contain occurrences of the same word in different contexts or senses. These texts can be analyzed automatically and word sense disambiguation can be employed to infer the context solely based on the contained words. For this purpose, pairwise similarities between single word occurrences can be calculated by masking and comparing word neighborhoods. Based on these similarities the occurrences can be clustered into potential contexts. This approach has been applied to a text corpus containing occurrences of the word "need".
coli_state (Details)
Text corpora contain occurrences of the same word in different contexts or senses. These texts can be analyzed automatically and word sense disambiguation can be employed to infer the context solely based on the contained words. For this purpose, pairwise similarities between single word occurrences can be calculated by masking and comparing word neighborhoods. Based on these similarities the occurrences can be clustered into potential contexts. This approach has been applied to a text corpus containing occurrences of the word "state".
coli_time (Details)
Text corpora contain occurrences of the same word in different contexts or senses. These texts can be analyzed automatically and word sense disambiguation can be employed to infer the context solely based on the contained words. For this purpose, pairwise similarities between single word occurrences can be calculated by masking and comparing word neighborhoods. Based on these similarities the occurrences can be clustered into potential contexts. This approach has been applied to a text corpus containing occurrences of the word "time".
 Hints:
Hints:
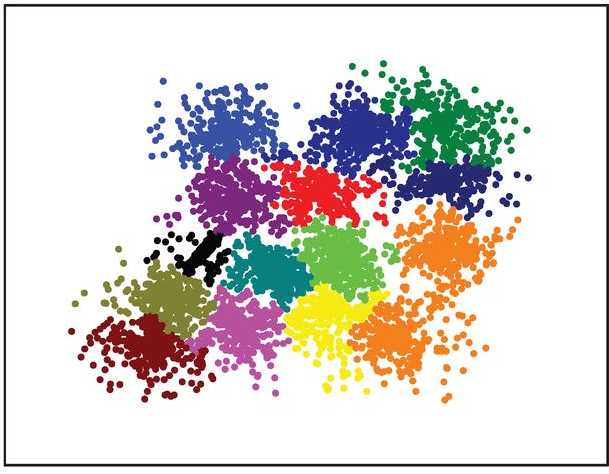 Synthetic 2-d data set with 5000 points in 15 Gaussian clusters with relatively high degree of cluster overlap.
Synthetic 2-d data set with 5000 points in 15 Gaussian clusters with relatively high degree of cluster overlap.